Literature Reading Workflow
Introduction
I have gone through many different iterations of how to keep track of my reading throughout my PhD and professional career. Here I document my latest version that so far has most of the kinks worked out, along with various improvements I would like to add in the future.
Towards the start of my PhD I begun tracking reading material and papers using Zotero, mainly because it seemed to have all the features I required and included browser plugins that would allow for easy adding of new papers that I came across. However over time Zotero felt more and more cumbersome to work with, especially when looking to extend the functionality to fit my own personal setup for reading and documenting my notes on given papers. This lead me to begin building out my own system for tracking papers and my notes related to them. Focussing on the Unix philosophy and trying to find small pieces of software that can be composed to be perfectly tailored to my own workflows.
Workflow
The aim of my reading workflow is to try and keep all of the relevant papers for a given research project together in a central database along with all my notes on the given papers. Recently it has also extended to put an Large Language Model (LLM) in the loop to allow for quick query answering if wanted. As yet the LLM in the loop aspect has not proven to be indispensable but has provided a small amount of value in specific usecases and will hopefully improve over time as LLMs improve.
Tracking Publication
For this aspect all I needed was to be able to easily add papers to a central store with some form of tagging. For this I found the pubs, this is a Command Line Interface (CLI) based tool for keeping track of publications. It follows the Unix philosophy of doing one thing and doing it well, this interface allows me to track publication whilst adding tags for various research projects the paper is relevant for. This also means for a given tag I can easily generate a bibtex file making referencing in future documentation easier.
I have a few bash scripts that allow for me to add documents to pubs by simply providing a web address, as shown below:
| |
This takes an input URL to a pdf and then adds the document to pubs, this allows me to have a keyboard shortcut setup in my browser to add any document to my library without the need for a custom browser plugin to be created. Note that it also creates an accompanying file in my notes with the bibtex information for the paper already added, this will become relevant later when looking at my reading workflow on laptop.
Reading & Understanding
Once the document has been added to the library Syncthing is used to sync the pdf files and the .pubs directory to my central server. This means that it is instantly available on all my devices, laptop (usually the place that I add the reference anyway), phone and my E-Reader.
The E-Reader is where most of my reading takes place, I resisted using E-Readers for awhile for reading research papers mainly due to the fact that black and white figures often made understanding results difficult. With the introduction of affordable colour eink I have now fully converted all my academic reading. The reading experience is pretty much as expected here, the documents automatically appear on the E-Reader thanks to Syncthing and I’m able to easily read and take notes using a stylus.
There is one integration I have made here however, and that is the introduction of an LLM in the loop. As shown below I am able to highlight a section of the document and attach a note with a question, this is then given to an LLM with the document in the prompt in order to ground the model. This processing is handled on the central server that contains powerful GPUs allowing for large-ish models to be run in order to answer the given questions, the document is then updated and the answer appears on the E-Reader as shown in Figure 1.

I also have it setup so that different highlight colour correspond to different LLM models, allowing for models trained on maths, code or other tasks to be used in order to get better results depending on the question. The model used for the inference is given in the note so I know what model and version was used to generate the answer. In my experience this functionality is hit and miss, most of the time the LLM answers the question at such a high-level it doesn’t add much insight, however when using task specific LLMs the usefulness improves greatly. There have been a few instances in using this setup where I have been impressed with the result and feel it actually aided in helping me understand the work quicker.
All of these annotations are automatically synced to my Laptop, so once having read through the paper initially on the E-Reader I am able to seamlessly move to the laptop if the paper is relevant to my current research. When opening the paper on my laptop three main windows are opened, the first is a Zathura instance of the paper opened to the last viewed page (left pane), my notes on the paper are then opened (top-right pane) and finally an LLM chat is opened with the content of the paper already prepared in a prompt for asking quick queries. This setup can be seen in the Figure 2.
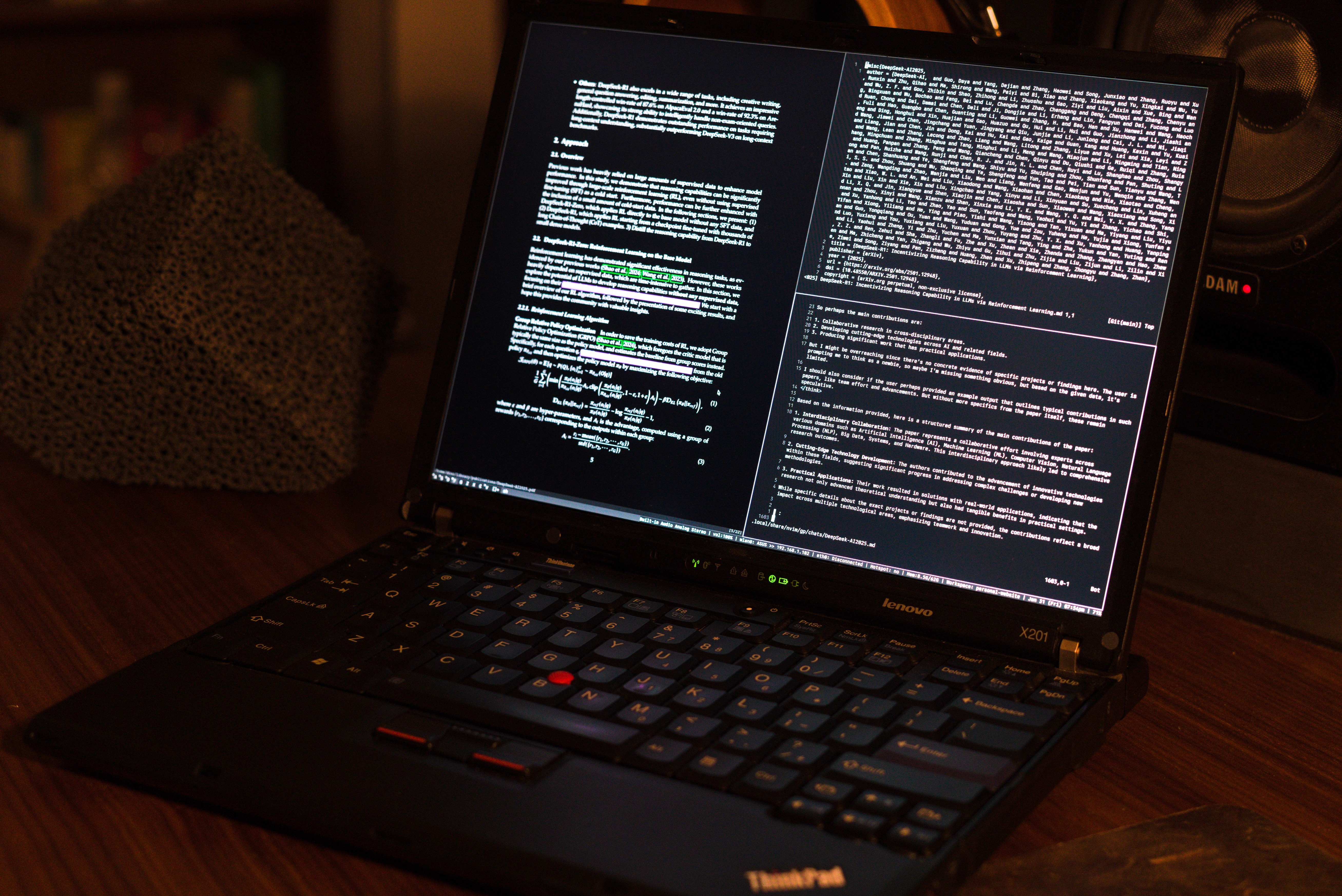
This allows me to easily catch up on my notes for the paper and see all of the annotations that I may have made on the E-Reader, along with any questions that have been highlighted and answered by an LLM. This is usually the stage in reading that I actually apply the contributions of the paper to the current research I am working on this I would add extra tags to the paper here in order to make sure it is tracked for the specific project.
Work to be done
I have covered the tracking and reading of papers in this post but have left out the actually referencing of the paper into written work. This is mainly because I have not implemented my ideas in this area yet. Previously with Zotero I would export a given library for a project as a bibtex and then manually add this to the latex document I was writing, this is currently the exact same setup I am doing here, just generating a bibtex file for a given tag from pubs and manually referencing the docs in my latex document. I however see an improvement to be made here where I can write a vim binding that will allow for searching through my entire library, or a subset of papers in order to reference them with the appropriate bibtex added to the document if needed and the correct \cite{} made in the document when a paper is selected.
I have so many small improvements to various projects I jump in and out of like this that who knows when I’ll get around to implementing the above ideas.